Abstract
Background: Relapse is a main event of therapy failure for patients with acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) undergoing allogeneic hematopoietic stem cell transplantation (alloSCT). Hence, a novel strategy to detect minimal residual disease (MRD) is required for earlier therapeutic intervention in impending post alloSCT relapse. Recent evidence indicated that screening of driver mutations by next generation sequencing (NGS) followed by detection of circulating tumor DNA (ctDNA) via droplet digital PCR (ddPCR) is a promising approach for MRD monitoring in patients with MDS (Paul Y, et al. Blood. 2017). Here, in this study, we investigated whether serial ctDNA monitoring could identify patients who are most likely to relapse after alloSCT.
Methods: We performed a retrospective study in 31 patients with AML and MDS undergoing alloSCT. Cell-free DNA (cfDNA) was extracted from serum samples. We subjected primary tumor DNA, extracted from a diagnostic peripheral blood or bone marrow (BM), and buccal swab DNA, to NGS, identifying one or two somatic driver mutations. We designed unique ddPCR assays for each somatic mutation identified (n=25). The primary endpoint was cumulative incidence of relapse rate (CIR) and the secondary endpoint was overall survival (OS). CIR and OS were calculated by the Kaplan-Meier method. The log-rank test was used for group comparison.
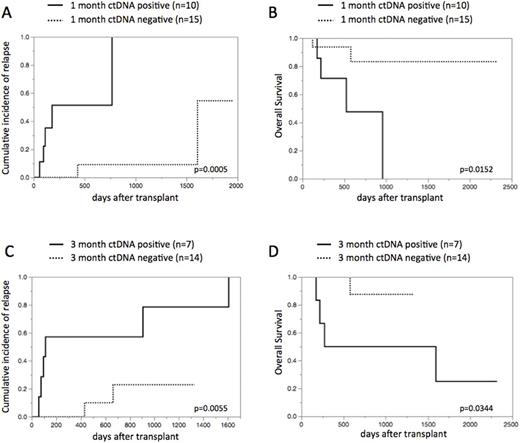
Results: 31 patients were enrolled in our study: our cohort consisted of 10 patients with de novo AML, 15 with AML with myelodysplasia related changes, and 6 with MDS. The median age was 52 years (17-68 years) and 5 patients (16%) are in complete remission at alloSCT. Stem cell source included related bone marrow (n=2) and cord blood (n=29). The preparative regimen included myeloablative conditioning (n=17) and reduced intensity conditioning (n=14). NGS screening identified 26 somatic driver mutations in 30 of 31 (97%) cases, with 2 mutations found in 5 individuals. Overall, we could construct ddPCR assays for 29 of 31 (94%) patients: these mutations included NRAS, TP53, NPM1, DNMT3A, GATA2, etc. NGS and ddPCR assays had a good concordance in assessing the same tumor rich samples with regard to the variant allele frequency (VAF).The amount of cfDNA at diagnosis was significantly higher compared with paired cfDNA at remission (n=23; p=0.0133, paired t-test). Of 37 matched serial time points, VAF between BM and serum ctDNA indicates apparent correlation (r2=0.92, p<0.0001). CtDNA was detected in 10 of 25 (40%) samples at 1 month after alloSCT and 7 of 21 (33%) samples at 3 months (termed "ctDNA1-positive" and "ctDNA3-positive", respectively).We next evaluated whether the positivity of ctDNA at 1 month and 3 months after alloSCT was associated with higher CIR and lower OS. As expected, patients with ctDNA1-positive and ctDNA3-positive had significantly superior CIR and inferior OS: 2-year CIR 51% in ctDNA1-positive patients vs. 9% in negative patients, p=0.0005 (Figure 1A); 2-year OS 48% in ctDNA1-positive patients vs. 83% in negative patients, p=0.0152 (Figure 1B); 2-year CIR 57% in ctDNA3-positive patients vs. 23% in negative patients, p=0.0055 (Figure 1C); 2-year OS 50% in ctDNA3-positive patients vs. 88% in negative patients, p=0.0344 (Figure 1D), respectively.
Conclusions: In summary, personalized ctDNA monitoring could detect MRD, which alloSCT has failed to eradicate, and thus identify patients with high-risk of relapse. This might allow for clinicians to initiating earlier therapeutic interventions for impending relapse. Further prospective studies in a much larger cohort of AML and MDS undergoing alloSCT are required to confirm these findings.
Figure 1. CIR and OS based on ctDNA status at 1 month and 3 months after alloSCT
Nagamura-Inoue: Tsubakimoto Chain CO.: Research Funding; AMED grant: Research Funding; Rohto Pharmaceutical Co., Ltd.: Research Funding. Tojo: AMED grant: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal